PEAKS AB 3.5 Software New Features
PEAKS AB automates protein de novo antibody sequencing using liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) datasets from orthogonal enzyme digests. From confident de novo sequence tags, the full protein sequence is assembled using a weighted de Brujin graph1.
In peaks AB 3.5, the software package is equipped with additional tools to enhance sequencing accuracy, PTM characterisation and result presentation including:
- Retention time prediction for isobaric differentiation.
- Improved intact mass deconvolution algorithm.
- lle/Leu differentiation by the signature w-ions included in sequence validation function.
- Added support for in-depth O-glycan analysis.
- Merged glycopeptides into peptide mapping view.
- Support for timsTOF data
- Improved UI and report.
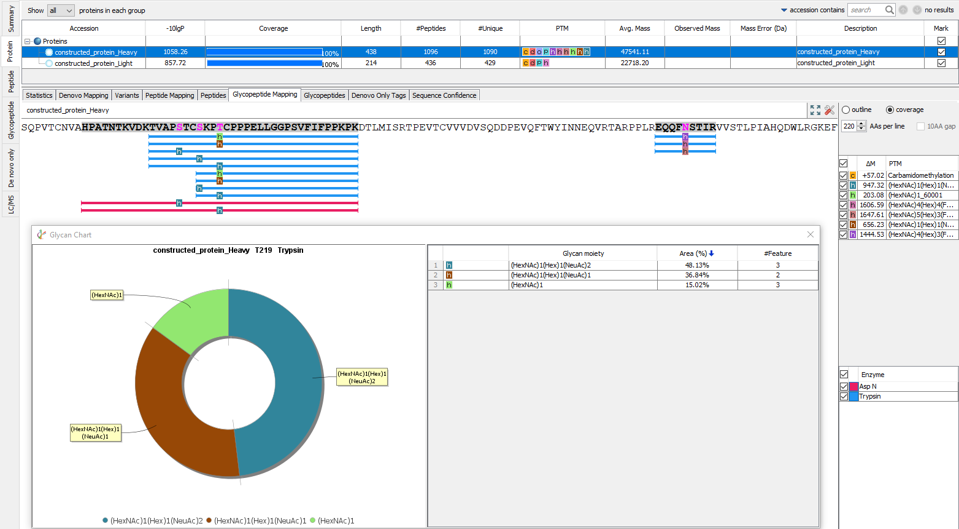
In-Depth Glycan Profiling
In our previous PEAKS AB build we introduced N-linked glycan profiling. In addition to this, PEAKS AB 3.5 now supports identification and profiling of O-linked glycans. This tool performs in-depth glycan profiling for both N- and O-linked sites identified in the heavy and/or light chains of the antibody. In addition to accurate glycopeptides mapping to the assembled antibody sequence, enzyme-based glycan profiling displays the composition and relative abundance of each glycan at a selected glycosylation site. Glycan composition and structure annotation are provided within each glycopeptide spectrum and are based on glycan fragment ions that match to an O-linked or N-linked glycan databases. Accurate localisation of the glycan at each site is achieved by identifying fragment ions of glycan moieties associated with the peptide backbone.
Retention Time Prediction to Differentiate Isobaric Residues
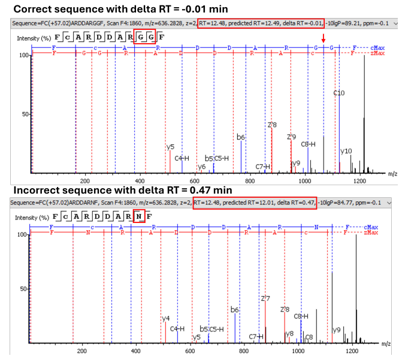
Introduced in PEAKS AB 3.5 is the prediction of retention time (RT). This function allows isobaric residues to be distinguished and ensures that peptide sequences used for protein assembly are accurate. Predicted RT and delta RT can be found in de novo table for further investigation.
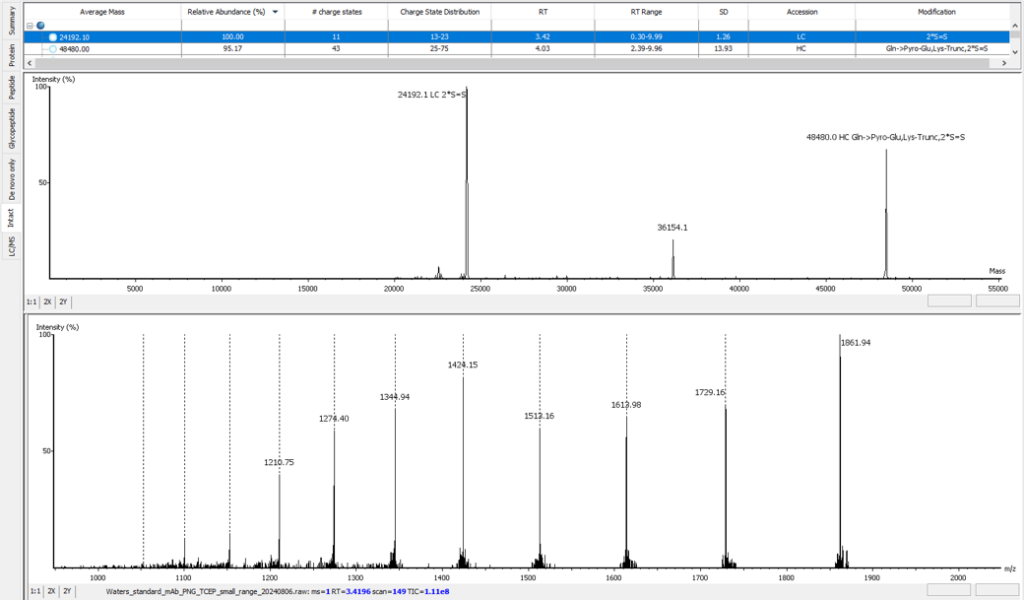
Advanced Intact Mass Deconvolution Analysis
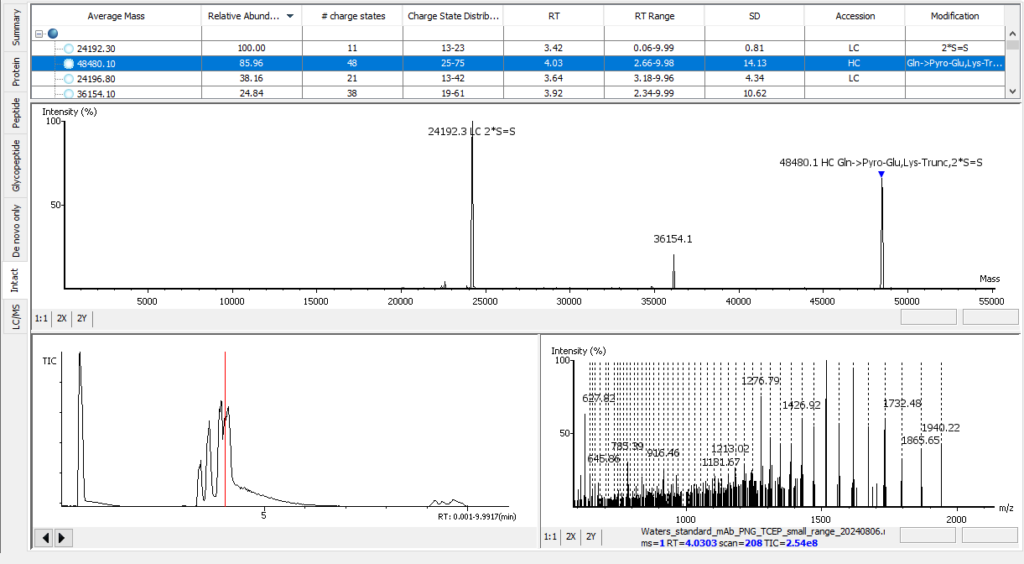
PEAKS AB 3.5 has an improved deconvolution algorithm which enables automated, efficient and accurate intact mass analysis. Intact mass measurements of proteins are performed to validate the assembled sequence and presence of any modifications such as N-terminal pyro-glutamate (Gln -> Pyro-Glu), C-terminal lysine truncation (1*Lys-Trunc) and N-linked glycosylation (`*A2G0F), as shown below. In addition, support from intact mass values have been integrated into the bottom-up peptide mapping result view.
Enhanced 3-Tier Ile/Leu Differentiation
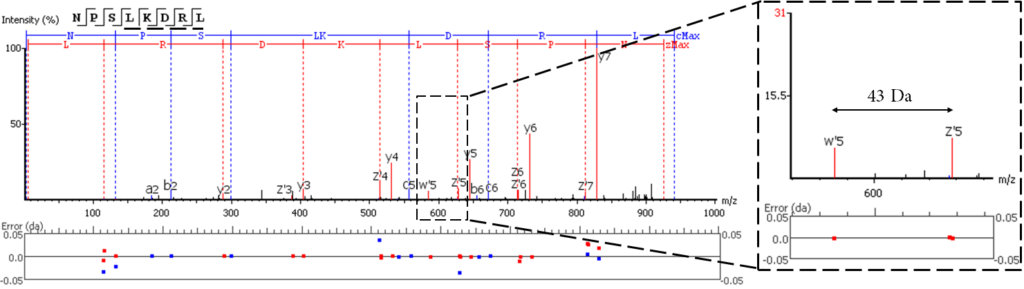
EThCD or EAD fragmentation produces signature w-ions from the characteristic loss of -C2H5 (-29 Da) from isoleucine and -C3H7 (-43 Da) from leucine. PEAKS AB uses this information, plus enzyme digestion specificity and homolog database analysis for improved Ile/Leu discrimination. New in PEAKS AB 3.5, Ile/Leu are differentiated in the Sequence Validation workflow, providing further confidence that the expected antibody sequence is correct.
- z ion with I on its N-terminus loses ethyl (-29)
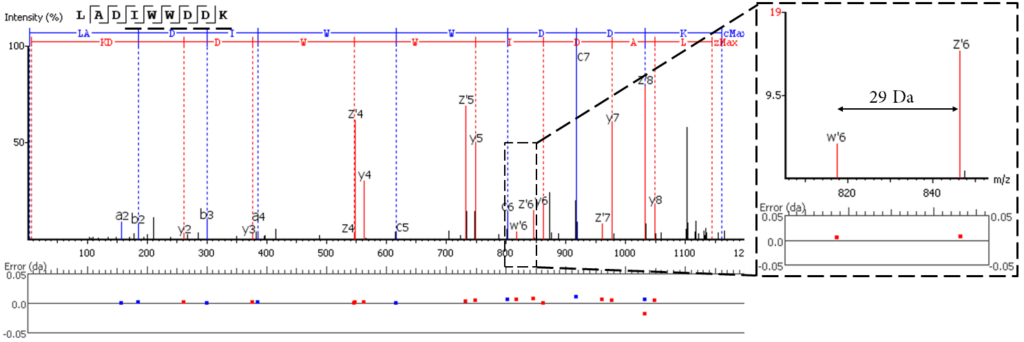
- z ion with L on its N-terminus loses isopropyl (-43)
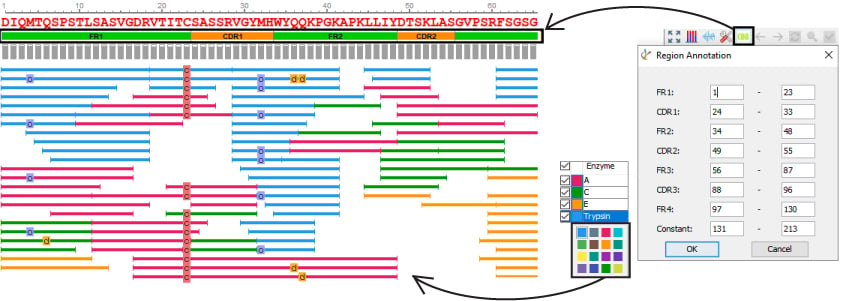
Greater Customisation Capabilities
PEAKS AB 3.5 now applies a local confidence (CAA) score for each PSM, and the selection of the final sequence path is based on this CAA score.
- Customise enzyme colours
- Automated and customisation CDR annotation
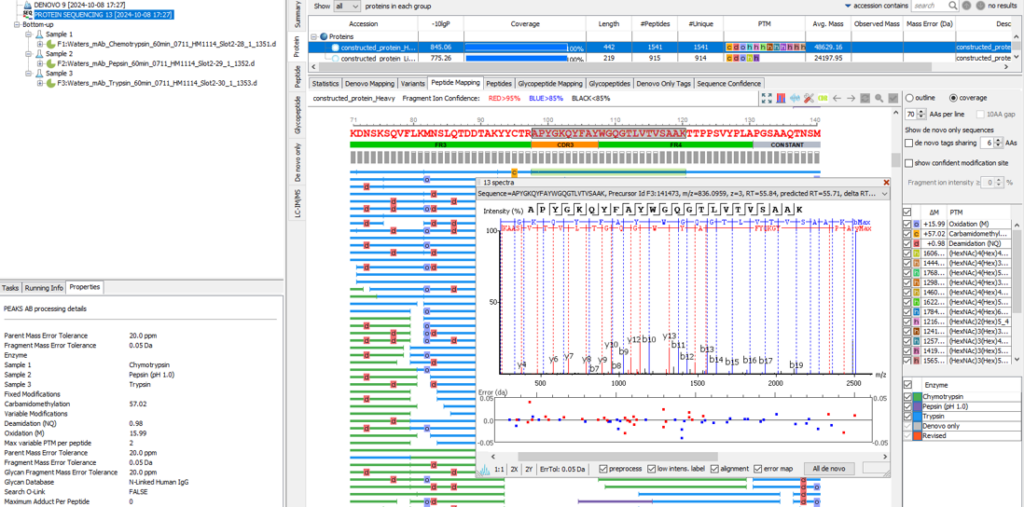
Support for timsTOF Data
In addition to Thermo and Sciex data, Bruker timsTOF data is now supported in PEAKS AB 3.5. The user can simply upload raw Bruker data as .d folders during project set up for de novo protein sequencing or sequence validation workflows.
References
- Tran, N.H. et al. Complete De Novo Assembly of Monoclonal Antibody Sequences. Scientific Reports. 6:31730. 26/08/2016.



