Bioinformatics Solutions Inc. (BSI) researchers have published their novel de novo sequencing algorithm in the recent Proceedings of the National Academy of Sciences (PNAS). The study successfully applies a novel deep neural network model to de novo peptide sequencing from tandem mass spectrometry (MS/MS) data. DeepNovo, the proposed de novo sequencing algorithm, achieves major improvement of sequencing accuracy over state-of-the-art methods and subsequently enables complete assembly of protein sequences without assisting databases.
DeepNovo was evaluated on a wide variety of species and considerably outperformed state-of-the-art methods, achieving 7.7–22.9% higher accuracy at the amino acid level and 38.1–64.0% higher accuracy at the peptide level. DeepNovo was further used to automatically reconstruct the complete sequences of antibody light and heavy chains of mouse, achieving 97.5–100% coverage and 97.2–99.5% accuracy, without assisting databases.
The computational model of DeepNovo is shown as below (more details please refer to the full article):
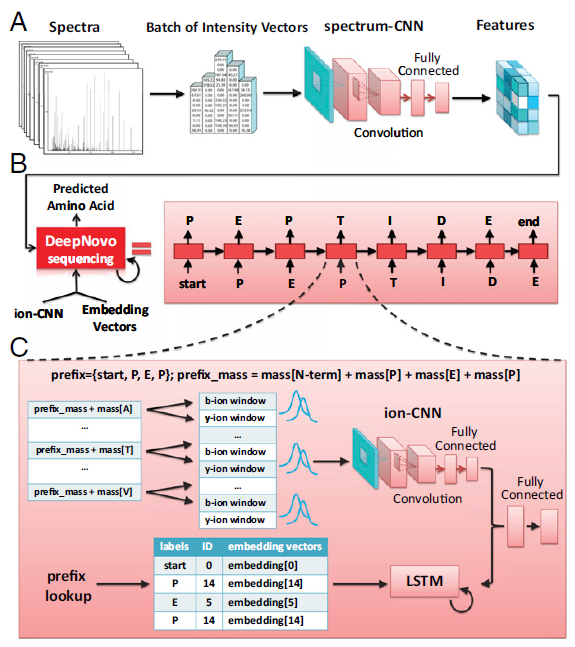
“The DeepNovo model for de novo peptide sequencing. (A) Spectra are processed by the CNN spectrum-CNN and then used to initialize the LSTM network. (B) DeepNovo sequences a peptide by predicting one amino acid at each iteration. Beginning with a special symbol start, the model predicts the next amino acid by conditioning on the input spectrum and the output of previous steps. The process stops if, in the current step, the model outputs the special symbol end. (C) Details of a sequencing step in DeepNovo. Two classification models, ion-CNN and LSTM, use the output of previous sequencing steps as a prefix to predict the next amino acid.” -Tran, N.H., et al.
The full article can be found here: Tran, N.H., et al. De novo peptide sequencing by deep learning. PNAS. 114(29). 18/7/2017.

